实现一个大而美的kernel往往是不现实的,在各种硬件上针对特定case下的kernel很容易能够突破针对通用case实现的kernel,所以实现的kernel在某些case下表现好,某些case下表现的就不是那么好,如何快速正确的找到各个kernel的性能曲线,既能更好的了解这个kernel的优势和不足,也能为后续kernel选择重要的参考。这里的这篇就是针对kernel选择而做的一个尝试。
这里做了两个工作,第一个是刷了一下之前写的两组conv在常见case下的各种时间数据,后面给出头部信息,第二个是用sklearn的一些Manifold方法尝试对这些数据进行分割,这里选了两种看上去效果不错的方法,在这里对比一下。
这里给了两个perf文件,如果只有一个就用一个就好了,注释掉第二个,csv的文件头如下, type我这里写的是gemm或者direct,这两个是自己实现的kernel,cudnn代表直接跑的cudnn的kernel:
1
| input_num,in_channels,out_channels,height,width,kernel_h,kernel_w,pad_h,pad_w,stride_h,stride_w,dilation_h,dilation_w,group,type,latency,
|
代码给出如下:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
| import csv
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import offsetbox
from sklearn.manifold import TSNE
from sklearn import (manifold, datasets, decomposition, ensemble, discriminant_analysis, random_projection)
from mpl_toolkits.mplot3d import Axes3D
self_perf = list()
cudnn_perf = list()
perf_file1 = "/Users/zhangshuai20/workspace/direct.csv"
with open(perf_file1, "rb") as csvfile:
spamreader = csv.reader(csvfile, delimiter=',')
for row in spamreader:
if row[14] == 'type':
continue
if row[14] != 'cudnn':
self_perf.append(row)
else:
cudnn_perf.append(row)
perf_file2 = "/Users/zhangshuai20/workspace/gemm.csv"
with open(perf_file2, "rb") as csvfile:
spamreader = csv.reader(csvfile, delimiter=',')
for row in spamreader:
if row[14] == 'type':
continue
if row[14] != 'cudnn':
self_perf.append(row)
else:
cudnn_perf.append(row)
feature_data = list()
self_label = list()
cudnn_label = list()
for row in self_perf:
row_list = list()
for r in row[:13]:
row_list.append(int(r))
feature_data.append(row_list)
self_label.append(float(row[15]))
for row in cudnn_perf:
cudnn_label.append(float(row[15]))
print('Computing t-SNE embedding')
hasher = ensemble.RandomTreesEmbedding(n_estimators=100, random_state=3, max_depth=3)
X_transformed = hasher.fit_transform(feature_data)
pca = decomposition.TruncatedSVD(n_components=2)
result = pca.fit_transform(X_transformed)
fig = plt.figure()
idx = 0
count = 0
ratio = 0
ax = plt.subplot(111)
for item in result:
if self_label[idx] < cudnn_label[idx]:
ax.scatter(item[0], item[1], marker='*', c = 'r')
ratio += self_label[idx] / cudnn_label[idx]
count += 1
else:
ax.scatter(item[0], item[1], marker='+', c = 'orange')
idx = idx + 1
plt.show()
print count
print "sass kernel is better than cudnn about: " + str(ratio / count)
print "better than cudnn % : " + str(float(count) / float(len(cudnn_label)))
|
我这里实现了两组不同的kernel,分别同cudnn进行对比,来指引我们什么时候选用什么样的kernel,红色是自己实现的kernel速度快于cudnn,黄色是cudnn的快于自己实现的kernel
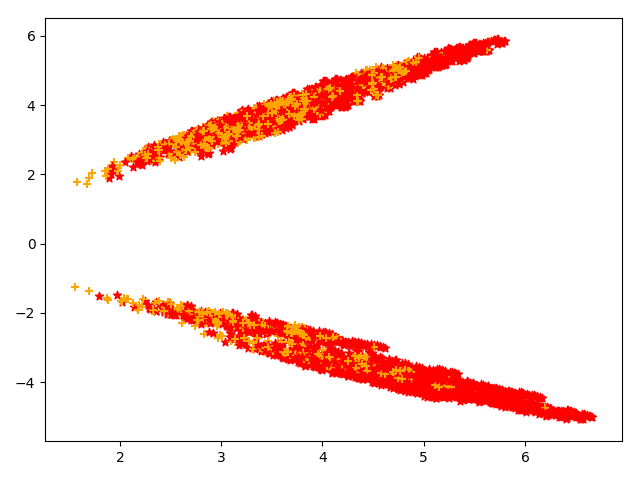
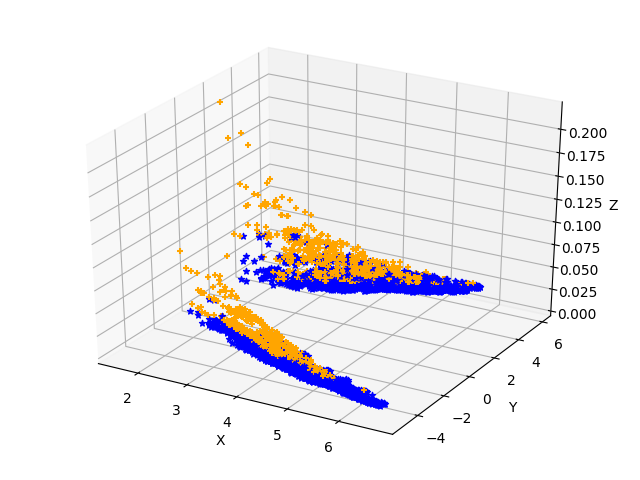
本文标题:找到kernel性能分界线的尝试
文章作者:throneclay
发布时间:2019-07-18
最后更新:2022-08-03
原始链接:http://blog.throneclay.top/2019/07/18/kernel_split/
版权声明:本博客所有文章除特别声明外,均采用 CC BY-NC-SA 3.0 CN 许可协议。转载请注明出处!

